Figure 1. Phylogenetic tree analysis of signal peptides (as of 01/02/2012, 1766 peptides with precursors).
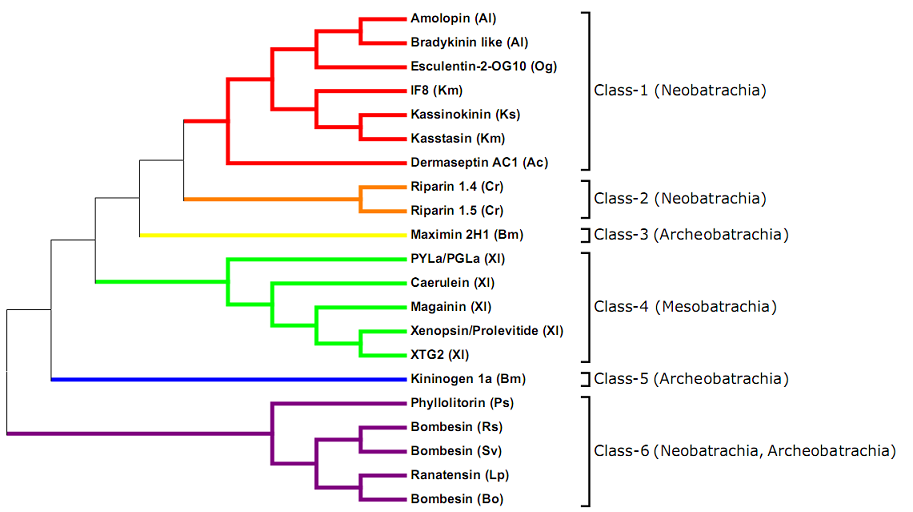
Topographic phylogenetic tree of representative signal peptides for anuran entries in the DADP. The p-distance* was calculated by the Neighbour joining method (pairwise deletion) using Molecular Evolutionary Genetics Analysis (MEGA) software1, version 5. Selected representative mature peptides for each type of signal sequences are indicated (Ac = Agalychnis callidryas; Al = Amolops loloensis; Bm = Bombina maxima; Bo = Bombina orientalis; Cr = Crinia riparia; Km = Kassina maculata; Ks = Kassina senegalensis; Lp = Lithobates pipiens; Og = Odorrana grahami; Ps = Phyllomedusa sauvagii; Rs = Rana shuchinae; Sv = Sanguirana varians; Xl = Xenopus laevis).
* - p-distance is the proportion (p) of amino acid sites at which the two sequences to be compared are different. It is obtained by dividing the number of amino acid differences by the total number of sites compared.
1 Tamura K, Peterson D, Peterson N, Stecher G, Nei M, and
Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using
maximum likelihood, evolutionary distance, and maximum parsimony
methods. Molecular Biology and Evolution 28, 2731-2739.
Signal peptide classes were determined by constructing a phylogenetic tree of different signal peptide types. Only peptide types that have more than three members were used. To determine which peptides would be the representative sequence, a consensus sequence was determined within each class. This was done by calculating the amino acid frequency for each position within the signal peptide and constructing a sequence that represents amino acids with the highest frequency for each site. A naturally occurring signal peptide was chosen that either was identical or most similar to the consensus, to serve as a representative sequence for this signal peptide class. For classes 1, 3, and 5 there were respectively 100, 152 and 1 signal peptides identical to the consensus. For classes 2, 4 and 6 this procedure was not carried out either because of a too small number of peptides (class-2) or because the peptides were too divergent in sequence and length (classes 4 and 6). In these classes all non-identical signal peptides were used for tree building. To divide them in classes, signal peptides were aligned by ClustalW and then a tree was constructed using MEGA 5 software. In the resulting tree (Figure 1) we identified six different clades that we named Classes 1-6. Some peptides present in the database have not entered into any of the mentioned classes and were named as unclassified peptides.
Class-1
It is the largest class by far and encompasses 1508 precursors or 85% of all precursors in the database. This class of signal peptides is found exclusively within the suborder of Neobatrachia and includes both antimicrobial and non-antimicrobial peptides. Most of the signal peptides in this class are 22 amino acids long and share a double lysine (KK) motif in the fifth and the sixth position as in Amolopin, representative of the consensus sequence for this class. Due to deletions, the KK doublet is in positions 4/5 and 2/3 in Brevinin-2TP1 and Odorranain-F1 precursors respectively. Also, substitutions of one of the two lysine residues are present in some signal peptides. The examples are arginine and asparagine in position five and glutamate and arginine in position six. Odorranain-D1 and IF-8-like peptide lack the SP KK motif altogether but share great similarity throughout the rest of the sequence with other members of this class. Kassinakinin and Kasstasin have the KK motif in position 3/4 and they have been found in only one kind of precursor in Kassina senegalensis and Kassina maculata respectively.
Alignment representing the most common features of Class-1 signal peptides:
| Amolopin | MFTLKKSLLLLFFLGTISLSLC-- | (identical to consensus sequence) |
| Brevinin-2TP1 | MFS-KKSLVVLFFLGTISLSLC-- | |
| Esculentin-2OG10 | MFTMRKSLLVLFFLGTISLSLC-- | |
| Dermaseptin-AC1 | MAFLNKSLLLVLFLGLVSLSIC-- | |
| Bradykinin-like | MFTLKESLLLLFFLGAISLSLC-- | |
| Odorranain-D1 | MFT----LLLLFFLGTISLSLC-- | |
| Odorranain-F1 | M---KKSLLVLFFLGIVSLSLC-- | |
| IF-8 | MLTLRTSMLLLFFLGMVSFSLA-- | |
| Kassinakinin | MM--KKSMLLLFFLGMVSLSLAYN | |
| Kasstasin | MM--KKSMLLLFFLGMVSFSL--- |
Red letters denote conserved amino acids.
When consensus query MFTLKKSLLLLFFLGTISLSLCEEEGDADE (consensus Class-1 SP extended with eight amino acids from the acidic propiece region typical for Brevinin-2E) is used with the HMMER homology tool http://hmmer.janelia.org/search, it gives 1446 hits, out of which 1441 are anuran host defense peptides (some are fragments excluded from DADP).
Class-2
This class contains four riparins, skin-derived bioactive peptides devoid of antimicrobial function from an Australian species Crinia riparia. All four signal peptides are identical with a single variation of valine/phenylalanine at the sixth position.
Sequence of Class-2 type:
| Riparins | MKIIV(V/F)LAVLMLVSA |
Class-3
This class is the second largest with 217 members. All these peptides belong to the same group of maximins, multifunctional peptides from the family of Bombinatoridae (Archeobatrachia). All signal peptides have conserved F3 and Y5.
Consensus sequence of Class-3 type peptide:
| Maximin | MNFKYIVAVSFLIASAYA |
Class-4
This class contains 11 peptides of four different types. All peptides come from Mesobatrachia species of Pipidae family (Xenopus sp.). Members of this class include both antimicrobial and non-antimicrobial peptides. Their common feature is a conserved lysine residue in the third position and a conserved cysteine in the eighth and fifteenth position with the exception of caerulein whose signal peptide has additional six residues at the C terminus and serine-15 instead of cysteine-15.
Sequences of Class-4 type:
| PYLa/PGLa | MYKQIFLCLIIAALCATIMA------ |
| Magainin | MFKGLFICSLIAVICANA-------- |
| Caerulein | MFKGILLCVLFAVLSANPLSQPEGFA |
| Xenopsin/Prolevitide/XTG | MYKGIFLCVLLAVICANSLA------ |
Class-5
This class contains eight peptides of the kininogen group, within the suborder of Archeobatrachia, family Bombinatoridae. These peptides are exclusively non-antimicrobial and their common feature is the tryptophan residue at the fourth position.
Consensus sequence of Class-5 type:
| Kininogen(Bombina) | MRLWFCLSFFIVLCLEHFTGTLA |
Class-6
This class contains eight members from suborders of Neobatrachia (Ranidae and Hylidae) and Archeobatrachia (Bombinatoridae). All peptides in this class have non-antimicrobial function. The common feature of these signal peptides is the proline residue in the fifth position.
Sequences of Class-6 type:
| Ranatensin | MTTIPAIGILPI-DFLTILLLFSFISHS--- |
| Bombesin (Bombina) | MSAIPLNRILPL-GFLFHLLIFSFISLSSC- |
| Bombesin (Rana) | MSLLPAVKVLPL-GYLGIVLVFSLILRSAMV |
| Phyllolitorin | MSAVPFTRVLLISGFLAHLLLSTFVTLTVC- |
Unclassified SPs:
These signal peptides (total of 10 entries) were categorized as unclassified since very few of them are present to form a proper class as they are too different from one another and any of the peptides in the other six classes. The number in parenthesis indicates the number of unique SPs of the given type:
| Cathelicidin(Al) | (1) | MGLSATLWFLMGVAAGSMAS | Neobatrachia |
| Brevinin-1SH(Ps) | (1) | MLFYLPISVSSSRRDA | Neobatrachia |
| Andersonin-A1(Oa) | (1) | MFLTPLKIKFLIPFPANLEKRP | Neobatrachia |
| Xenoxin-1(Xl) | (1) | MRYAIVFFLVCVITLGEA | Mesobatrachia |
| Pleiotrophin(Xl) | (1) | MRHQHGLFMLALLAFLFVITVLG | Mesobatrachia |
| Midkines(Xt,Xl) | (2) | MELRAFCVILLITFLAVSSQA | Mesobatrachia |
Al- Amolops loloensis; Ps- Pelophylax saharicus; Oa-Odorrana andersonii; Xl- Xenopus laevis; Xt- Xenopus tropicalis